 AdaFilter explicitly guarantee replicability of scientific findings from multiple studies / individuals
AdaFilter explicitly guarantee replicability of scientific findings from multiple studies / individualsAbstract
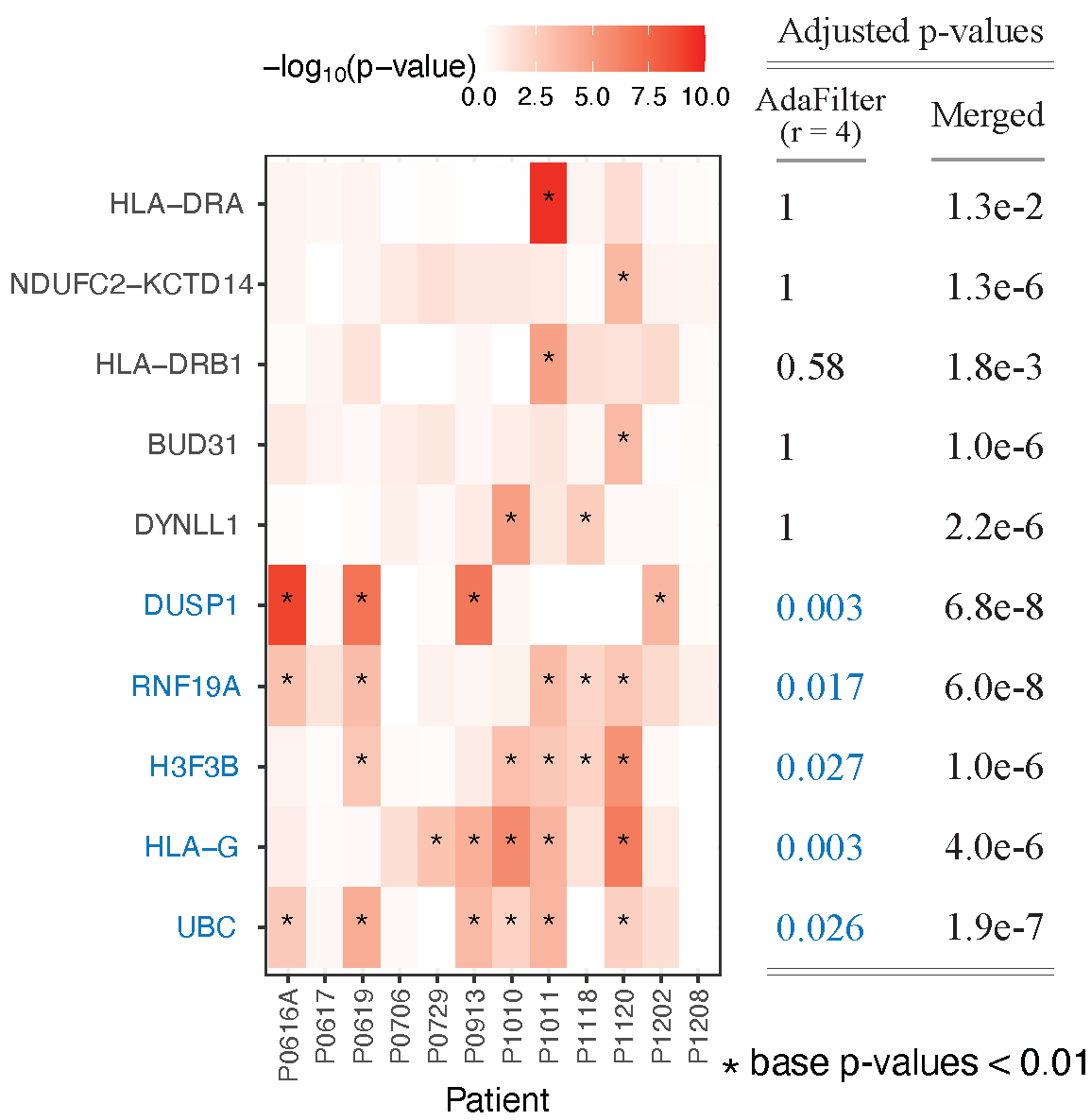
Replicability is a fundamental quality of scientific discoveries: we are interested in those signals that are detectable in different laboratories, different populations, across time etc. Unlike meta-analysis which accounts for experimental variability but does not guarantee replicability, testing a partial conjunction (PC) null aims specifically to identify the signals that are discovered in multiple studies. In many contemporary applications, e.g., comparing multiple high-throughput genetic experiments, a large number M of PC nulls need to be tested simultaneously, calling for a multiple comparisons correction. However, standard multiple testing adjustments on the M PC p-values can be severely conservative, especially when M is large and the signals are sparse. We introduce AdaFilter, a new multiple testing procedure that increases power by adaptively filtering out unlikely candidates of PC nulls. We prove that AdaFilter can control FWER and FDR as long as data across studies are independent, and has much higher power than other existing methods. We illustrate the application of AdaFilter with three examples: microarray studies of Duchenne muscular dystrophy, single-cell RNA sequencing of T cells in lung cancer tumors and GWAS for metabolomics.